AmplifX 1.00 : first "final" release
Sunday 9th January 2005.Thanks to the suggestions and remarks of many users of the 0.9x versions I could improve AmplifX at a point it justify the version number of 1.00 !
New features or corrections
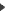 New calculation of quality taking of account TM (bases
stacking method), the %GC, the presence of polyX and the presence of
dimers.
New calculation of quality taking of account TM (bases
stacking method), the %GC, the presence of polyX and the presence of
dimers.
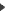 New window drawer containing of the details on the primer quality
and a field "observations" in order to associate any type
of data text to each primer.
New window drawer containing of the details on the primer quality
and a field "observations" in order to associate any type
of data text to each primer.
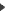 The different matches from the same primer at several places on the sequence targets are now easily visualized by the simultaneous change of their color when the mouse flies over them.
The different matches from the same primer at several places on the sequence targets are now easily visualized by the simultaneous change of their color when the mouse flies over them.
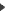 Taking into account of "U" (or u) in the targets and oligos - > automatically transformed into T (or t)
Taking into account of "U" (or u) in the targets and oligos - > automatically transformed into T (or t)
 Correction for the importation of text files of primers created with textEdit
for example (nonconventional character of end of line for Mac)
Correction for the importation of text files of primers created with textEdit
for example (nonconventional character of end of line for Mac)
 Translation of "contaminant" words in French in the English version (thank you to Todd Geders to have raised them and have proposed a translation)
Translation of "contaminant" words in French in the English version (thank you to Todd Geders to have raised them and have proposed a translation)
 New table for the calculation of the score of annealing of the oligos: allows to better balance strong matching primers and weak matching ones.
New table for the calculation of the score of annealing of the oligos: allows to better balance strong matching primers and weak matching ones.
 New and more readable drawings of the matches in two forms : strong (score > 170) and weak ones ; the same for amplified sequences drawings
New and more readable drawings of the matches in two forms : strong (score > 170) and weak ones ; the same for amplified sequences drawings
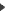 Primer or amplified fragment whose information tab are visualized are painted in a special color
Primer or amplified fragment whose information tab are visualized are painted in a special color
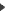 Choice is given to show weak matches a primer can get only if it have strong matches with the sequence or to show all weak matches.
Choice is given to show weak matches a primer can get only if it have strong matches with the sequence or to show all weak matches.
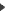 the columns "Length" and "Quality" of the list of primers are sortable by numerical size, the other columns remain sortable by alphanumeric order (Think of using string inputs of type Pr0001, Pr0002,... in field ID for example).
the columns "Length" and "Quality" of the list of primers are sortable by numerical size, the other columns remain sortable by alphanumeric order (Think of using string inputs of type Pr0001, Pr0002,... in field ID for example).
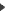 The menu " open a sequence" is working :one can open sequence files in different text format like GenBank, EMBL,FastA, GCG. The other text formats being regarded as "rough" format i.e. all found « ATCGU »characters are being imported
The menu " open a sequence" is working :one can open sequence files in different text format like GenBank, EMBL,FastA, GCG. The other text formats being regarded as "rough" format i.e. all found « ATCGU »characters are being imported
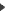 Improvement of the possibility of design the primers directly in AmplifX thanks to a live quality calculation while selecting nucleotides in the field "Target" (TM and global note of quality)
Improvement of the possibility of design the primers directly in AmplifX thanks to a live quality calculation while selecting nucleotides in the field "Target" (TM and global note of quality)
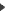 « Select All » menu works with field « info »
« Select All » menu works with field « info »
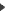 Horizontal scroll bar to change the visible area of the graphic area when it is zoomed in
Horizontal scroll bar to change the visible area of the graphic area when it is zoomed in
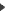 New menus : « Upper cases » and « Lower cases » to change the style of the current selection of the field « Target » (useful to visualize some parts into the graphic view and copy to another application)
New menus : « Upper cases » and « Lower cases » to change the style of the current selection of the field « Target » (useful to visualize some parts into the graphic view and copy to another application)
Here is the list of all the nice guys that encourage me by their constructive remarks :
Jean-Paul Herman (my boss in my real job !), Jean-Louis Franc, Denis Becquet, Andrea Cabibbo, Christophe Hennequin, Corentin Cras-Méneur, Immo Junghärtchen, Jean-Paul Boissel, Markus Winter, Patrick Trieu-Cuot, Todd Geders.